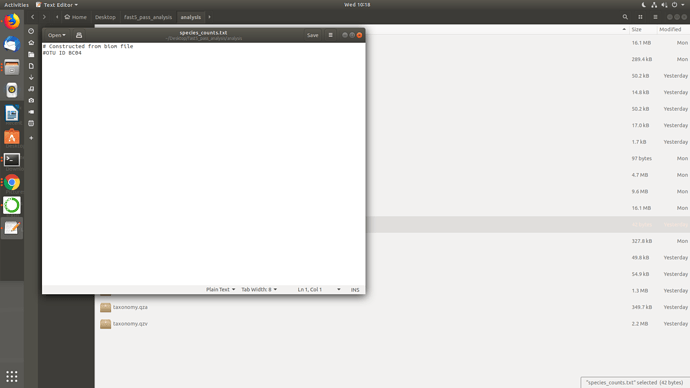
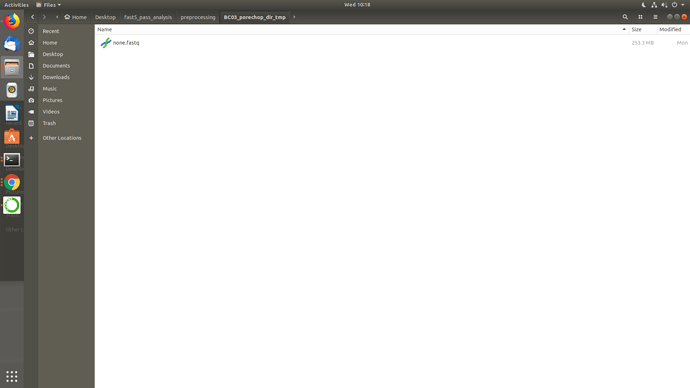
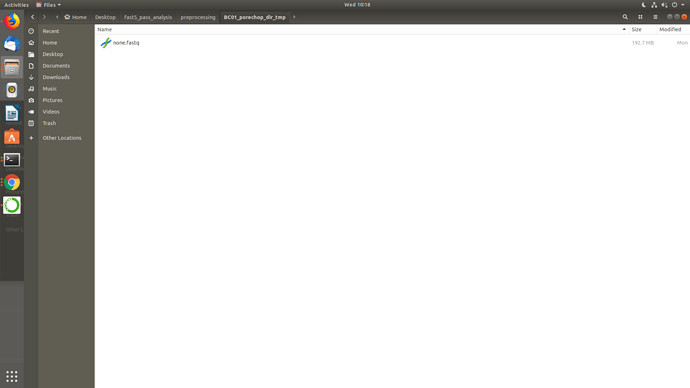
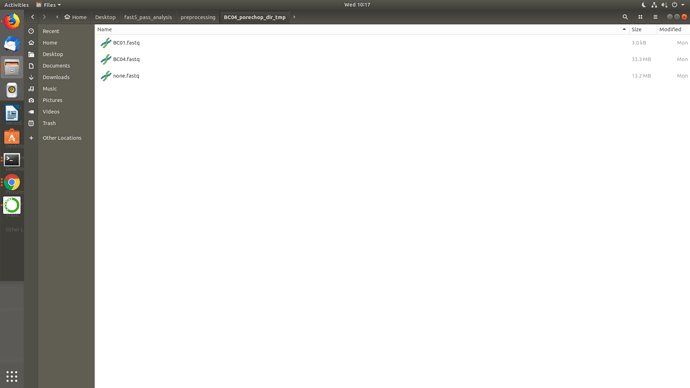
Well, I uninstalled and reinstalled porechop manually in that conda env, checked the paths then ran Launch_MinION_mobile_lab.sh and it worked but didn't go full pipeline and the temp directories seems really odd BC01, BC03 came as none.fastq while BC04 came okay and also BC01 in BC04.. I am not getting it.. I am sharing screenshots.
I also ran the ./MetONTIIME.sh but it also ended before completion. Can you please take a look !!!
Thanks
Hi @uddalok06
I just saw this seems to be a known Porechop issue. Basically, Porechop finds an ex-aequo identity value for adapters in forward and in reverse orientation, and is not able to decide the correct orientation of adapters. Since the demultiplexing by Guppy seems to be working, as shown by pycoQC pie chart, this issue should be solved by skipping the second round of demultiplexing by Porechop. In particular, you should:
git clone GitHub - MaestSi/MetONTIIME: A Meta-barcoding pipeline for analysing ONT data in QIIME2 framework
cd MetONTIIME
chmod 755 *
and then modify the values in the config file as you previously did. This time you also have to set disable_porechop_demu_flag <- 1, which by default is set to 0. I think this time BC02 could be included too in the analysis. You don't need to reinstall all the software and recreate the MetONTIIME_env conda environment, as the only modifications are in the config_MinION_mobile_lab.R and in the MinION_mobile_lab.R files.
I hope this helps, let me know.
Thanks a lot @MaestSi it seems MetONTIIME works fine for my Nanopore data, went through the full pipeline and it it worked. Thanks for the discussion.
Hi Simone,
Don't you think that 99 quite high given the error model of ONT, would something closer to 90 not be more compatible with the data (or am I off with the meaning of 99?)
Thanks
Welcome to the forum @splaisan!
I have not looked over @MaestSi's pipeline in detail, but as far as I can tell I think the answer is no, 99 is not too high.
Here 99% OTU clustering is performed on the database, NOT the query sequences. The reason for this is to dereplicate the sequences while still preserving a reasonably high level of resolution between individual species.
This is distinct from OTU clustering on query sequences, where the goal is instead to dereplicate and reduce sequence noise by collapsing low-abundant sequences into more abundant similar sequences.
So the 99% OTU clustering of the reference database is a whole other thing entirely — it is not nanopore data, and should be clustered at a very high % similarity to retain more information. Just because the reference sequences are clustered at 99% does not mean you can't perform OTU clustering at a lower % similarity if that is your preference.
I hope that helps!
Hi, @splaisan,
sorry for the late reply. As for your question, I think that @Nicholas_Bokulich, which is much more experienced than me on microbiome analysis, already provided a very complete and detailed answer, and I totally agree. Regarding nanopore data analysis in EPI2ME 16S (and MetONTIIME) I think the goal is to maximize the number of sequences assigned at species level, since the default database we are using is not clustered at all, we are not using a stringent identity threshold for assigning taxonomy (set at 77%) and we are retrieving only up to 1 top hit for each read. However, you can increase the confidence for your assignments looking at higher taxonomic levels, as genus or family, for whom taxonomic classification with Nanopore reads is reported to work well. Using QIIME2 default parameters would retrieve up to 10 top hits and then assign the read to a "consensus taxonomy", namely the least common ancestor of the retrieved top hits. Increasing the minimum percent identity or increasing the number of retrieved top hits for consensus taxonomy assignment may work as well for retrieving only "high-confidence" species, but I have not tested the impact of these parameters on the percent of unassigned reads and on the reliability of identified species yet.
thanks Nicholas for the detailed answer, I am trying to digest the info now ![]() and definitely need to read through the full QIIME before I can get the global picture.
and definitely need to read through the full QIIME before I can get the global picture.
Thanks Simone for your comments.