Hello everyone,
I am analyzing a COI amplicon dataset and running into a taxonomy mismatch issue after filtering out unwanted lineages. The error appears specifically when I try to collapse my feature table at the genus level.
I first applied taxonomy-based filtering to remove unwanted groups (mitochondria, chloroplast, fungi, bacteria, archaea, plants, etc.):
qiime taxa filter-table \
--i-table "$INPUT_TABLE" \
--i-taxonomy "$INPUT_TAXONOMY" \
--p-exclude "Fungi,fungi,Bacteria,bacteria,Archaea,archaea,chloroplast,Chloroplast,Carnivora,Streptophyta,Chlorophyta,Mucoromycota,Ailurus,Aves,Bacillariophyta,Unassigned" \
--o-filtered-table "$FILTERED_TABLE"
qiime taxa filter-seqs \
--i-sequences "$INPUT_REP_SEQS" \
--i-taxonomy "$INPUT_TAXONOMY" \
--p-exclude "Fungi,fungi,Bacteria,bacteria,Archaea,archaea,chloroplast,Chloroplast,Carnivora,Streptophyta,Chlorophyta,Mucoromycota,Ailurus,Aves,Bacillariophyta,Unassigned" \
--o-filtered-sequences "$FILTERED_SEQS"
I then attempted to collapse the table at the genus level:
qiime taxa collapse \
--i-table "$FILTERED_TABLE" \
--i-taxonomy "$INPUT_TAXONOMY" \
--p-level 6 \
--o-collapsed-table "$OUTPUT_DIR/collapsed-table-genus.qza"
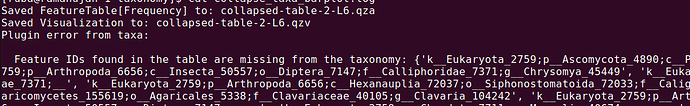
The issue
When collapsing at level 6 (genus), I get the following error:
How to solve this error.
Is it okay to continue with this error