Hello!
This is my first post to the forum and, though it seems like a well-documented issue, I’ve had trouble finding a solution on my own using the previous topics and would greatly appreciate you all’s expertise and assistance.
I’ve been working with a sequence dataset of 16S paired-end sequences (V4 region using 515F & 926R primers) derived from fish gut contents using Qiime2 version 2024.5.0 installed on a server using conda. I received the sequences untrimmed from the sequencing center and trimmed them using cutadapt with the following command:
qiime cutadapt trim-paired
--i-demultiplexed-sequences demux_fishgut_16s.qza
--p-front-f GTGYCAGCMGCCGCGGTAA
--p-front-r CCGYCAATTYMTTTRAGTTT
--p-discard-untrimmed
--p-match-adapter-wildcards
--p-match-read-wildcards
--p-cores 25
--o-trimmed-sequences demux_edna_16s_cutadapt.qza
--verbose
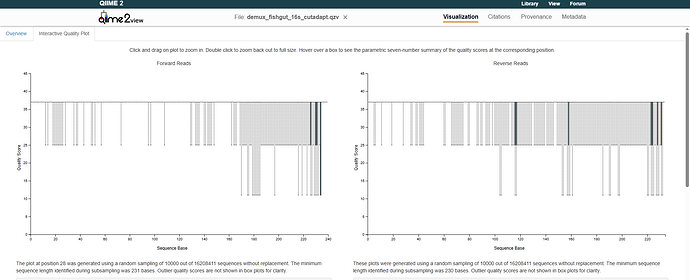
I received the following quality plot when viewing the output:
From here, I attempted to denoise and merge with DADA2, but have found that, though I have a good amount of sequences passing the filter, very few are merging. The first command I ran was like so:
qiime dada2 denoise-paired
--i-demultiplexed-seqs demux_fishgut_16s_cutadapt.qza
--p-trunc-len-f 230
--p-trunc-len-r 220
--p-trim-left-f 0
--p-trim-left-r 0
--p-n-threads 16
--o-denoising-stats dns_fishgut_16s_230_220
--o-table table-fishgut-16s-230-220
--o-representative-sequences rep-seqs-fishgut-16s-230-220
--verbose
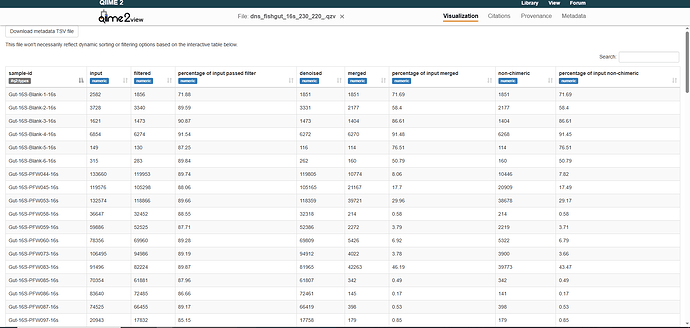
From there, I received about 24-25 samples out of 210 with >50% of reads merging, with a large proportion under 5% merging. An example table can be seen here:
I’ve tried a number of different truncation settings including 230F/180R, 230F/230R, 220F/200R, and I haven’t had much different results with each. I read a few other topics and am suspicious that this may be a sign of significant host amplification? Though I’m not entirely sure how feasible that is with fish gut samples… Either way, are there any folks who wouldn’t mind pitching in ideas for how I could improve merging for these samples? Our aim is high taxonomic resolution for community metabarcoding, so if that would be best served by continuing downstream analysis with single-strand reads, or trying a different denoising strategy, we’d be happy to look into it. I’d be very grateful for any of suggestions!