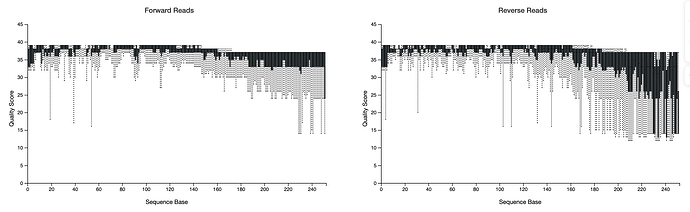
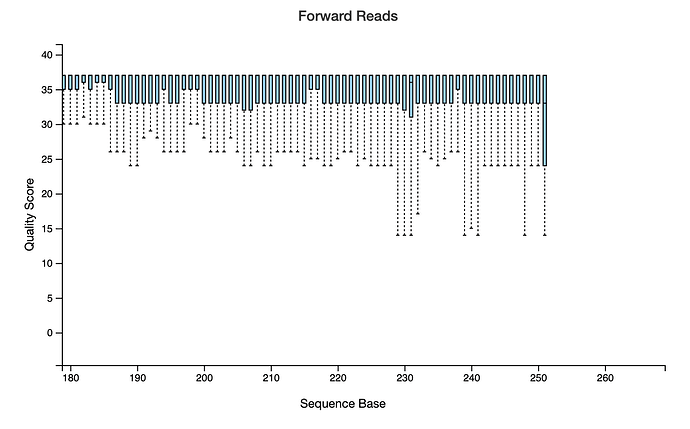
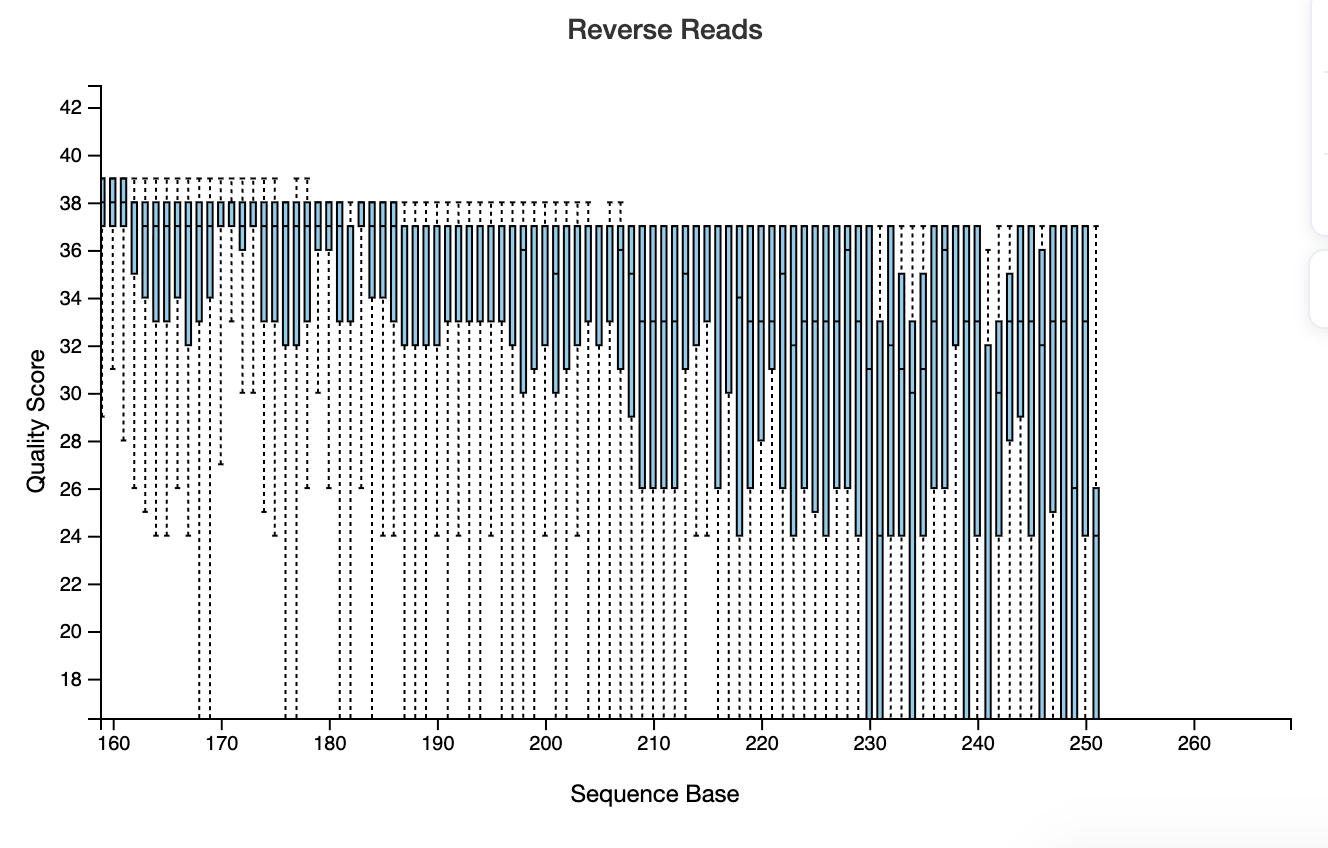
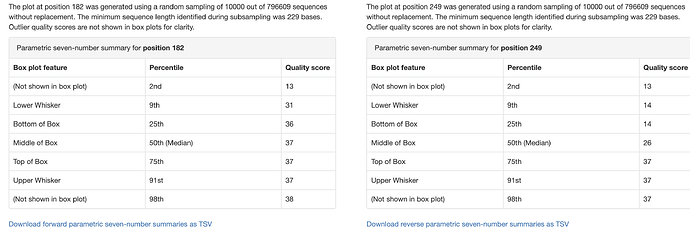
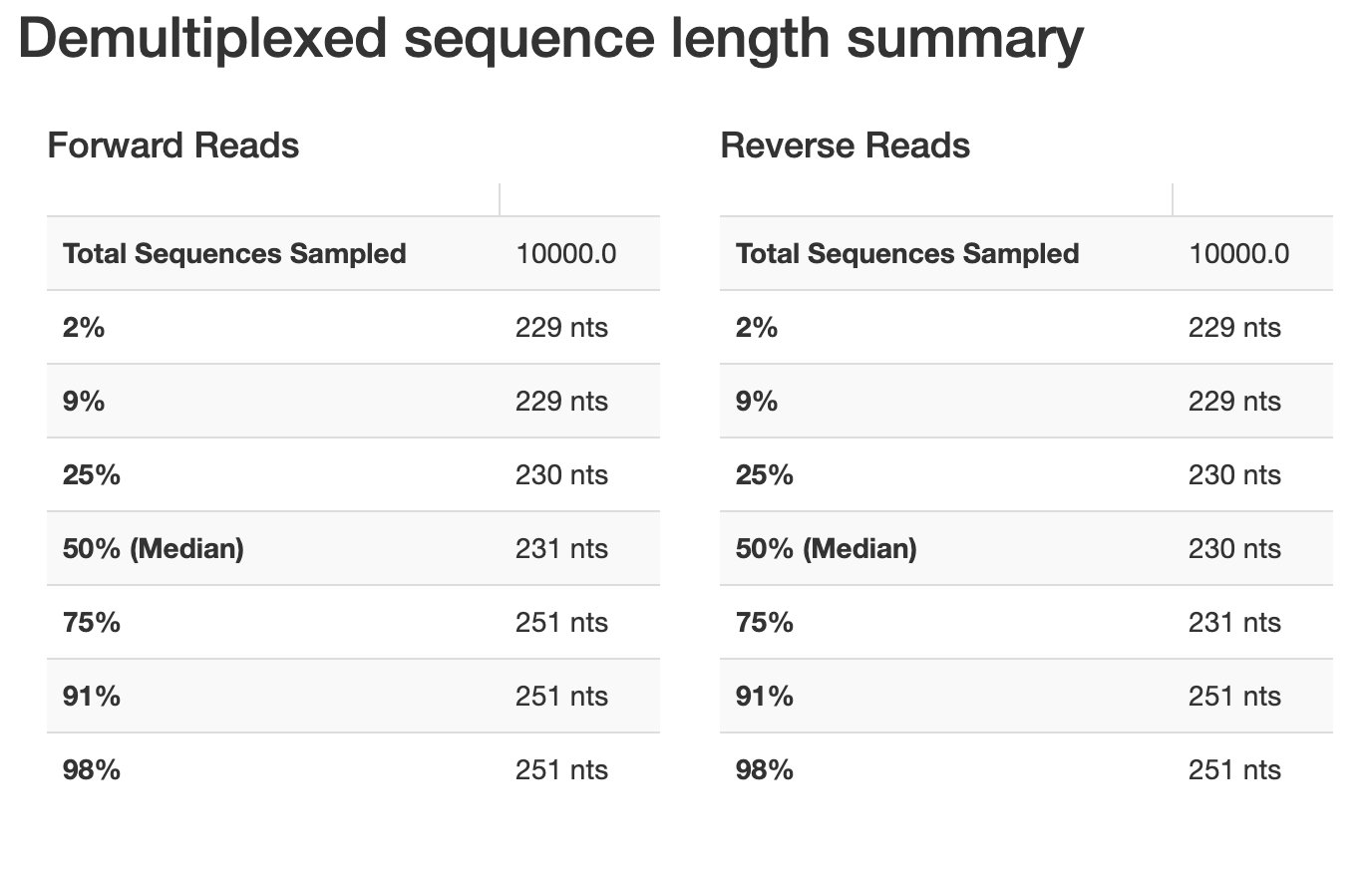
hi, below you can find my demux.qzv showing the read quality before DADA2 denoising, I cannot adjust my truncation values properly. I am replicating one of the studies done before and I know the OTU count that they had, yet mine results after all is almost half of it.
How can I adjust my parameters ?
I set this previously.
TRUNC_F=250 TRUNC_R=230 , qiime dada2 denoise-paired \\
--i-demultiplexed-seqs demux.qza \\
--p-trim-left-f 0 \\
--p-trim-left-r 0 \\
--p-trunc-len-f ${TRUNC_F} \\
--p-trunc-len-r ${TRUNC_R} \\
--o-table table.qza \\
--o-representative-sequences rep-seqs.qza \\
--o-denoising-stats denoising-stats.qza \\
--p-n-threads ${CPUS_PER_TASK} --verbose
demux.qzv (312.7 KB)