I don’t know if I can bring this up here, I’m new using qiime environment and working with taxonomic data.
I am working with 18S data and trying to train my classifier using PR2 database and assign taxonomy to my data but experiencing difficulty. Here is the script I used
qiime tools import
--type 'FeatureData[Sequence]'
--input-path pr2_version_5.1.0_SSU_taxo_long.fasta
--output-path pr2-seqs.qza
qiime tools import
--type 'FeatureData[Taxonomy]'
--input-format HeaderlessTSVTaxonomyFormat
--input-path pr2_taxonomy_fixed.tsv
--output-path pr2-taxonomy.qza
qiime feature-classifier extract-reads
--i-sequences pr2-seqs.qza
--p-f-primer CAAACGATGACACCCATGAA
--p-r-primer CCCCCTGAGACTGTAACCTC
--p-trunc-len 0
--o-reads kine-ref-seqs.qza
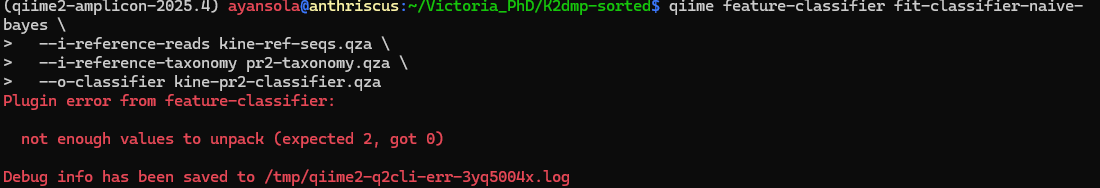
qiime feature-classifier fit-classifier-naive-bayes
--i-reference-reads kine-ref-seqs.qza
--i-reference-taxonomy pr2-taxonomy.qza
--o-classifier kine-pr2-classifier.qza
The error message
If I may ask what database is best to use to train 16S (bacteria), 18S (Apicomplexa and Kinetoplastida) and 28S (nematoda) classifiers?
Thanks