Hello,
I imagine that this is a basic question that I missed. How is it possible that we get singleton reads, for example, given that PCR would amplify each gene millions of times?
Thanks!
Hello,
I imagine that this is a basic question that I missed. How is it possible that we get singleton reads, for example, given that PCR would amplify each gene millions of times?
Thanks!
Hello @riVeRNAr,
Welcome to the forums! ![]()
Good question!
While PCR amplifies targeted regions many times, the absolute mass of nucleic acid after PCR is still very small. It's also often heavily skewed towards the most abundant amplicons, so the rare amplicons might only appear a dozen times across a MiSeq run. When these 10s of reads are spread out across 100s of samples, they could appear in a sample only once.
One other way we can get singletons it through PCR errors. While we are interested in biological variation, we also get "technical variation" when the polymerase makes a mistake on a base. This can result in a 'false' amplicon that appears 10s of times that is just one basepare different from it's 'true' amplicon that appears 1000s of times. Before we had better ways of denoising, singletons were sometimes removed because they were presumed to be errors of real amplicons.
Let me know if that helps,
Colin
Hey Colin! Thanks for your help here.
I get that PCR introduces errors, and I can understand why abundant sequences might be amplified more than rare sequences. I'm still unclear about the process, I hope it's okay that I ask more questions in a different way.
Let's say I have one 16S DNA strand to start with, and I amplify that sequence for 20 cycles in a PCR machine. My understanding is that that would produce 1 million double stranded DNA molecules. How many reads would be produced from an Illumina MiSeq? My intuitive understanding is that the sequencing process would produce one read (one 16S region from one organism), and I'm unclear as to how. How could the same process produce one read from one sequence that, let's say, was not amplified during the PCR process, as I think you describe?
Hello again,
Of course!
Yep. 1 read after 20 cycles = 1* 2^20 = 1,048,576 = 1.48 million reads amplicons.
Yes, that one read would end up stuck to the MiSeq flow cell, when it would then undergo bridge amplification forming a cluster of on the flow cell that is large enough to be visible during sequencing-by-synthesis.
Illumina has a video about it here. It's got a loud intro and lots of background music, but it's pretty helpful. https://www.youtube.com/watch?v=HMyCqWhwB8E
The bridge amplification step is done with PCR, so if an error is made in the first PCR cycle, that same error would be amplified leading to a low quality or inaccurate base call in the final read.
So in a perfect world, 1 amplicon makes 1 cluster on the flow cell makes 1 read in the fastq file. And even in a perfect world, errors can propagate through this process.
There's a lot to discuss in this process, and I'm still not sure if I'm answering your full question... is this making any sense? ![]()
Colin
Thanks, Colin. I'm getting a clearer picture of the sequencing process. Though, I'd like to go further.
Thanks for the video link, I ran into this before. I assumed the last ~40 seconds are talking about the bioinformatics pipeline, and not some Illumina sequence alignments that happen prior to creating a fastq file. Is that correct?
What happened to the other 1,048,575 reads that went into the sequencer from that one organism? This is where I'm still hung up.
Have a great weekend!
P.S. is read synonymous with amplicon?
Let's go!
That's right. The Illumina sequencing pipeline does include a computational photography step to convert the images from the flow cell into text files listing nucleic acids (fastq or bcl).
The steps they shows as "Data Analysis," I would describe as "Downstream Analysis" because it's after Illumina sequencing.
Yeah, we should probably break up the PCR step from the sequencing step.
That one nucleic acid sequence was primed and amplified 2^20 times during PCR, along with all the other regions from all the other organisms. This makes millions of amplicons from your millions of individual organisms! Even so, the total mass of PCR product, a.k.a your amplicons, is very small. There is just enough to appear as a hazy band, even after it's been stained with some crazy bright dye:
So if all of your amplicons / PCR products are just barely visible at the macroscale, how can we read the nucleic acid sequence of a single amplicon?
For the amplicons that do make it into the sequencer, many never stick to the flow cell, or they don't clonally amplify enough to be visible as a little dot.
Illumina is trying to image something impossibly small, and all these amplification steps, which seem like overkill, are just barely enough to produce a little blip of light on a glass slide.
If you are interested, here's what that slide really looks like:
The left shows one full lane of the flow cell.
The right shows 9 zoomed-in sections where you can actually see nucleic acid clusters where the Adenine has been added as a little glowing dot. (Source)
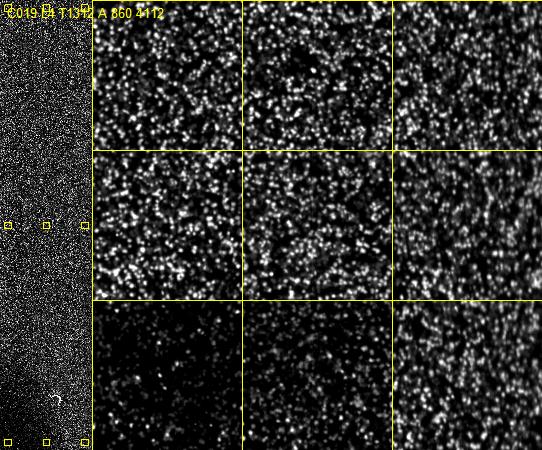
At the risk of stating the obvious, Illumina's fancy "fluorescently labeled reversible terminator" just lets them perform a normal PCR reaction one nucleic acid at a time.
They just added an 'A', and snapped a photo. ![]()
And it's still so small!
Colin
This is all really helpful! Thanks for engaging me on this topic, Colin. I think I get it.
I have a better understanding of how the sequencer works. I think I understand PCR. Though everything is more complicated than my mental representation.
Do we know what proportion of the PCR product sticks to the flow cell and is amplified enough to be visible? Just from my back-of-the-envelope calculation, it seems like a fraction of a percent (0.004%) for my average sample (30,000 reads/200 ASVs/4 16S regions per bacteria/1 million amplicons per 16S gene*100). Is that about right? That seems really small.
I suppose that, if the PCR solution is 100% homogenous, then what sticks to the flow cell could be accurately represented by the relative abundance of reads. That doesn't seem like a safe assumption to me based on my calculation. What are your thoughts?
I'm glad this has been helpful to you. I had fun writing it!
Illumina probably does, but I don't. It's also possible that the problem is somewhere else in the process; perhaps most of the reads anneal to the flow cell, but most of them don't clonally amplify. ![]()
Yeah.
There's a trade off here. More clusters sounds like a good thing, but then they overlap and you can't tell them apart. Illumina has a whole guide about avoiding 'over clustering' (pdf).
And it's not necessarily a problem because:
Exactly! Well said! ![]()
Colin
PS. I totally forgot to answer this
P.S. is read synonymous with amplicon?
You could also import un-amplified shotgun reads for analysis with q2-shogun, but in your study you have amplicon reads, so in this case they are synonymous.