Happy New Year QIIME 2 Forum! ![]()
If you're like me, you may be getting back to work this week and trying to remember where you left off with your analyses. To help with this, I built a little utility that, when provided with a specific Result (i.e., an Artifact or Visualization; referred to here as the target), and a directory to search (referred to here as the search directory), will locate all Artifacts in the target's provenance that are in the search directory. It will report back to you with the full path to each of the Results that it found, and list the Results that it didn't find (because they are not in the search directory).
The problem
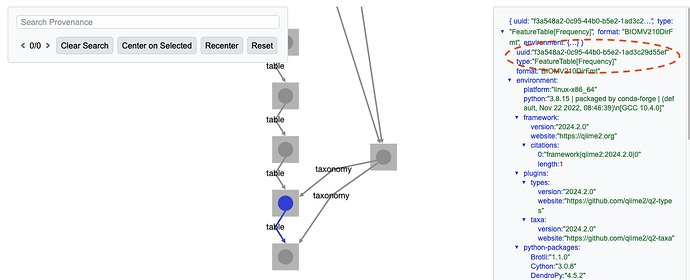
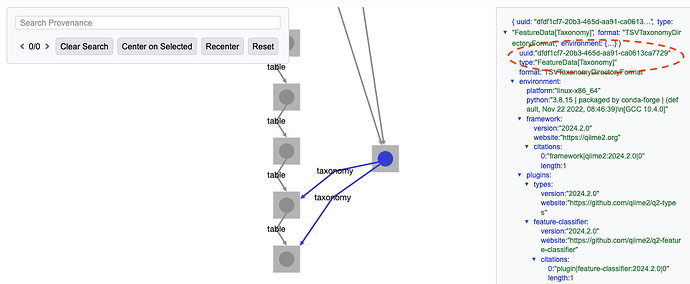
For example, imagine you're working with the full gut-to-soil data set, as available on Zenodo at Upcycling Human Excrement: The Gut Microbiome to Soil Microbiome Axis (supporting data). [1] You're looking at the full taxonomy barplot[2], and want to find the feature table and taxonomy that was used to generate it in the repository so you can recreate it with the new taxonomy barplot visualizer, qiime taxa barplot2, introduced in QIIME 2 2025.10. You can find the relevant Artifact UUIDs in the visualization's data provenance (see Figure 1 and Figure 2), but linking those UUIDs to filepaths on your system can take a while if you have a lot of .qza files.[3]
Figure 1: The barplot's data provenance shows that the
FeatureTable[Frequency] used to generate it had the UUID f3a548a2-0c95-44b0-b5e2-1ad3c29d55ef.
Figure 2: The barplot's data provenance shows that the FeatureData[Taxonomy] used to generate it had the UUID dfdf1cf7-20b3-465d-aa91-ca0613ca7729.
Enter artifinder!
Using Aritifinder's prov command (short for "provenance search"), you can provide the search directory and the path to the target result and get a list of paths to all Artifact's in the target's data provenance.
$ artifinder prov \
gut-to-soil-qiime2/ \
taxa-bar-plots-gtdb-r214.1-weighted-stool-taxonomy.qzv
The following is the output of running this, but I'll split it apart to talk about the components of the output. First, there is status and summary information printed to the terminal. This tells you how many .qza and .qzv files were found in the search directory. Then it tells you how many Artifacts are in the target's data provenance, and of those how many were found and not found in the search directory, respectively. If you want to see less of this information, you can provide the --no-verbose parameter when calling artifinder prov.
`artifinder` version: 0.0.1+3.ge78dfb5.dirty
Scanning search path for .qza and .qzv files...
Found 352 `Results` in search directory.
Parsing target's provenance...
Found 63 `Results` in target's provenance (not including target).
* 21 were found in the search directory.
* 42 were not found in the search directory.
Target `Result`:
38860088-ba62-4515-8de6-bd37550f561a Visualization taxa-bar-plots-gtdb-r214.1-weighted-stool-taxonomy.qzv
Then, for each Artifact that was found, it displays its UUID, its Artifact Class, and then the full path to where it can be found.[4] You can look for the UUIDs that we identified above in this list and we now know exactly what files on our file system were used as input. Voilà! ![]()
Found `Results`:
74a9e284-768c-4b0c-b2a1-d524885584bf FeatureData[Sequence] gut-to-soil-qiime2/dada2-output/hec-run1/asv-seqs.qza
7fb9bcf8-8dab-4d36-879b-45cbe4f1b8ca FeatureData[Sequence] gut-to-soil-qiime2/dada2-output/hec-run2/asv-seqs.qza
595d26ab-f0f0-44a0-a74e-94a88f7f858e FeatureData[Sequence] gut-to-soil-qiime2/combined/asv-seqs.qza
fd7dd96b-c401-4b17-95e5-8dcf34f1573f FeatureData[Sequence] gut-to-soil-qiime2/dada2-output/emp-soils/asv-seqs.qza
0ecd2151-a54b-4e2c-96b2-226e41a2a50b FeatureData[Sequence] gut-to-soil-qiime2/dada2-output/hec-run3/feature-table.qza
716fb24d-0077-435d-a3d1-f349f6d98312 FeatureData[Sequence] gut-to-soil-qiime2/dada2-output/hec-run4/asv-seqs.qza
dfdf1cf7-20b3-465d-aa91-ca0613ca7729 FeatureData[Taxonomy] gut-to-soil-qiime2/combined/taxonomic-annotations/gtdb-r214.1-weighted-stool-taxonomy.qza
e2bcbaf0-7c5c-4aa1-924e-53163d844265 FeatureTable[Frequency] gut-to-soil-qiime2/dada2-output/hec-run1/feature-table.qza
d90937f7-edaa-423d-858a-58c88cac53f9 FeatureTable[Frequency] gut-to-soil-qiime2/dada2-output/hec-run3/asv-seqs.qza
e07cd30e-a83c-4520-9092-02ab199a59ef FeatureTable[Frequency] gut-to-soil-qiime2/dada2-output/emp-soils/feature-table.qza
00d23d5b-20a1-4170-8619-89e5d3a679b4 FeatureTable[Frequency] gut-to-soil-qiime2/dada2-output/hec-run2/feature-table.qza
f3a548a2-0c95-44b0-b5e2-1ad3c29d55ef FeatureTable[Frequency] gut-to-soil-qiime2/combined/asv-table.qza
f522eb7d-412f-4dc7-a6ad-d267f7e308a4 FeatureTable[Frequency] gut-to-soil-qiime2/dada2-output/hec-run4/feature-table.qza
da396632-c2d8-4c1e-9237-e46f1a2f0c30 SampleData[PairedEndSequencesWithQuality] gut-to-soil-qiime2/demux/emp500/qiita-study-13114/88540/demux.qza
e80b5598-5432-4ac5-ba85-e46360065d64 SampleData[PairedEndSequencesWithQuality] gut-to-soil-qiime2/demux/emp500/qiita-study-13114/104886/demux.qza
3df45781-511d-4722-9773-cb1b546b59cc SampleData[PairedEndSequencesWithQuality] gut-to-soil-qiime2/demux/emp500/qiita-study-13114/104885/demux.qza
313811fe-08ad-4689-9117-971199f3ddef SampleData[PairedEndSequencesWithQuality] gut-to-soil-qiime2/demux/nano3/demux-nano-r3.qza
efa37f5b-0dfb-4939-bbd4-8be3b38c3eeb SampleData[PairedEndSequencesWithQuality] gut-to-soil-qiime2/demux/emp500/qiita-study-13114/104884/demux.qza
43514fc4-48e9-40b4-a5f3-4e530a4aaddb SampleData[PairedEndSequencesWithQuality] gut-to-soil-qiime2/demux/emp500/qiita-study-13114/104887/demux.qza
4bc12a54-a6db-480d-97bc-ed00face3c63 SampleData[PairedEndSequencesWithQuality] gut-to-soil-qiime2/demux/emp500/qiita-study-13114/104883/demux.qza
1ae3f1ba-6106-4063-a482-563183b079ee SampleData[PairedEndSequencesWithQuality] gut-to-soil-qiime2/demux/emp500/qiita-study-13114/88539/demux.qza
Finally, for the 42 Artifacts that were not found in the search directory, the UUIDs and artifact classes are listed. This, for example, lets you know that the TaxonomicClassifiers used in generation of this figure don't seem to be included in the search directory. If you don't want to see this information, you can provide the --no-include-missing parameter when calling artifinder prov.
`Results` not found:
1cf47da5-1538-4151-bc3a-f7c1af0cf424 EMPPairedEndSequences
2db99c5c-4899-46e5-b19a-ddce6ac40c3e EMPPairedEndSequences
85d9d87e-361f-4a92-87da-a43845da2c14 EMPPairedEndSequences
47094f5d-fdb7-4b1e-9e71-233531c95deb EMPPairedEndSequences
5b90b946-9ddb-4315-9c8f-83623cbda0b2 EMPPairedEndSequences
35009e42-86ca-4b92-926b-f3f561d1249f EMPPairedEndSequences
548ec3f4-3d90-4eb7-810e-2ee8d6eb52bf EMPPairedEndSequences
4afd1f08-72e8-4bbf-a73a-8c2bccb0b69f FeatureData[Sequence]
8162ad56-eb06-43d7-a52c-a96997d3098e FeatureData[Sequence]
6764be53-7936-431e-8cbc-5c293ad1f102 FeatureData[Sequence]
d775572a-79be-461f-95c1-586d5b1b6b13 FeatureData[Sequence]
b92a2676-3786-494c-9661-5edbf0934f42 FeatureData[Sequence]
745163cf-18b1-42e3-8a55-5a3acf569b5a FeatureData[Sequence]
3bfdf3fb-a031-4089-8be6-304c75c2b461 FeatureData[Sequence]
f3bbdad4-6274-431f-a56a-bfcbc8330a8b FeatureData[Sequence]
62e9def0-3461-436a-a101-c0fefba658a8 FeatureData[Sequence]
7fca1ee9-b6f3-43e8-9ca0-2f78c8bfc248 FeatureData[Sequence]
19fd8247-6c24-4e82-9753-c27ffcd7fb4d FeatureData[Taxonomy]
1bb2e87f-9fba-4f0e-acd1-11e12d66b1cc FeatureData[Taxonomy]
91a354b4-a7b0-43ca-9ad6-96857ae631e3 FeatureTable[Frequency]
f971ac95-9756-4e6f-b9b4-4a90740ad59a FeatureTable[Frequency]
dcd66f24-4f16-49a3-a679-cfe02ae81afd FeatureTable[Frequency]
9884b8bc-a061-42a3-a476-0646f17edf00 FeatureTable[Frequency]
4437b72f-b113-4809-b521-e9a1e0bc478a FeatureTable[Frequency]
b997d6da-ea6d-4d8f-b766-d5098534555e FeatureTable[Frequency]
cd182b04-46a0-4467-9974-84f7784060e1 FeatureTable[Frequency]
abd3b941-3c43-4107-8b20-128845f18a07 FeatureTable[Frequency]
89e06f0b-3471-454c-9ba6-7f56c0841bbd FeatureTable[Frequency]
03133bb2-10a3-42ab-b6db-5421e26f10a3 FeatureTable[Frequency]
dc57a837-7658-4f13-8edf-eb2f8063497a FeatureTable[Frequency]
08f3a02c-e2ff-4651-bf53-eda40beab872 FeatureTable[Frequency]
9cbcb60d-bac5-487a-8f48-56a79ee82851 FeatureTable[Frequency]
44f0f4ff-c425-474f-ad60-681693687044 FeatureTable[Frequency]
7a4309ce-f8ce-4f25-8b42-84d0294524c7 FeatureTable[Frequency]
c2e1f374-fa37-4ca4-9a7b-29bca6e6a359 FeatureTable[Frequency]
84fcf26d-3259-4886-bf0f-9718c393dff7 FeatureTable[Frequency]
f6ee7ac9-99b1-4dc5-a710-0a34171a9c27 FeatureTable[RelativeFrequency]
f8c447e0-75e1-4fc1-b984-637fcb726a57 SampleData[PairedEndSequencesWithQuality]
2b5b1192-8817-4ef2-9c81-292a6c58fe72 SampleData[PairedEndSequencesWithQuality]
301a6791-eaac-41e5-ab38-ba257b91ef5f SampleData[PairedEndSequencesWithQuality]
419d5c8a-253a-4cd5-83e8-61e1b276afc8 TaxonomicClassifier
771f54ec-fca4-45f0-b5f5-ad1cc0b03599 TaxonomicClassifier
Conclusion
I wrote this script recently because I had to re-run an ANCOMBC command that I originally ran a couple of years ago with a slightly different formula, and I wanted find the input feature table that I had previously used to make sure I was matching the previous analysis exactly (with the exception of the formula). This saved me a lot of time poking around trying to figure out where the specific files I worked with previously were. I'm sharing this with the hope that it will save you some time too.
If you'd like to try it out, you can find the installation instructions in the project's README. I have lots of ideas for other features that could be included, and there are some existing known limitations (see the project's issue tracker), so consider this to be alpha software. If there are things that would help you, please feel free to share.
Enjoy!
You can follow along with this if you like, but the dataset is around 16 GB so is a big download. Instead, it might be worth just reading and then trying it on some of your own data. ↩︎
This visualization is large - it'll take a couple of minutes to load using this link. ↩︎
You might be wondering why rachis doesn't store the input filepath(s) in data provenance. It's because filenames and locations can change, so neither are stable identifiers. An Artifact's UUID however is guaranteed to be unique and to not change, so unlike a filename or filepath, it is a stable identifier. ↩︎
In the output here, I edited these to be relative to the search directory so I'm not sharing information about my file system. ↩︎