Is it possible to determine the numbers species (or unique ASVs) belonging to a given phyla or family in each sample? e.g. how many species of Firmicutes are in a given sample. Thanks.
If you make a taxonomy bar plot, you can download the data as a .csv file and see those numbers.
Try that and let us know what you find! ![]()
EDIT: I don't think that answered you question! See discussion below
Hi Colin
I have 100 samples belonging to 30 sub-classes and 5 classes. I trying to generate box plots showing mean, max, min number of unique ASV in Firmicutes (and other phyla, families) in each class and subclass. I am currently using taxa bar plots to determine the number of unique ASV in each sample at Phylum level. I sort, split the table into each phylum, move the data to GraphPad Prism to generate the box-plots. This manual processing is manageable at phylum level, as there are 7 phyla (as of now). It becomes too much work at family level.
I get observed-features in a sample as part of core-metrics data. I was wondering if there is a script which can generate observed features per phylum, per family in a sample.
Another approach I am thinking of is filter based on taxonomy (phylum, family) and then run core-metrics analysis on each filtered table. This will also involve lots of steps.
Regards
I am not aware of any method to do it in Qiime2, only with custom scripts... ![]()
What scripting languages are you familiar with? Would you like me to put something together using R and Phyloseq?
I am familiar with R, qiime2R and phyloseq. I was able to create phyloseq object using qiime2R. Couldn't get done much beyond this. I was able to perform most of the analysis in qiime2. I would appreciate the scripts for phyloseq, if it is not too much work.
I'll see what I can mock up, starting with a phyloseq object as the input.
No promises! ![]()
Does this look like what you were looking for?
(We can adjust it so it shows exactly what you are looking for! ![]() )
)
Here's the code:
# Melt phyloseq object into data frame, with
# each row being a single feature.
ps.melt <- ps %>% psmelt()
# Now we can group by SampleType and count total number of Taxa in each
ps.melt %>%
group_by(SampleType, Phylum) %>%
summarise(nFeatures = n()) %>%
filter(nFeatures > 1000) %>%
ggplot(aes(x = SampleType, y = nFeatures, color = Phylum)) +
scale_y_log10() +
geom_point()
Looks like this will work. I will try this and update you. Thank you very much for doing this.
Hi Colin
I tried the script. I think it is returning number of reads in each phylum. See figure below.
See below my calculations in Excel.
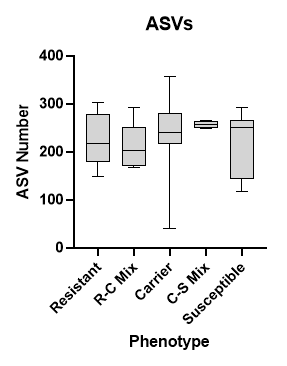
E.g. in Resistant group range of ASVs is 150-300. We have to count the number of unique ASV (or species, Genera) in a Phylum (or Family) in a group (or subgroup).
I appreciate your help, but do not want to take too much of your time. Thanks.
Thanks for trying that out. Looks like I missed a couple of important things.
First, after melting, make sure you are only counting features with abundance > 0.
Second, summarize these by Sample / SampleID as well.
Note: My data.frame column names may be different than yours, so be sure to update this code to match your 'melted' phyloseq object.
ps.melt %>%
# Remove ASVs that do not appear in a sample
filter(Abundance > 0) %>%
# For each Sample (with it's associated Type), take the Phylum...
group_by(Sample, SampleType, Phylum) %>%
# ... and count it
summarize(nFeatures = n()) %>%
# Filter for Phyla of interest (or don't and see all of them)
filter(Phylum %in% c("Firmicutes", "Actinobacteria")) %>%
# Plot
ggplot(aes(x = SampleType, y = nFeatures)) +
facet_grid(~Phylum) +
scale_y_log10() +
geom_boxplot()
Let's see if we can get our number to match at the Phylum level, then extend this to Family!
Thank you very much. It worked.
Could you please direct me to a tutorial about how to format the graph. I want to change axis labels, size, etc.
Yes! I built that using ggplot2. It's a pretty popular package, so you can google questions about formatting and find lots of examples, too!
This topic was automatically closed 31 days after the last reply. New replies are no longer allowed.


